Experience
Postdoctoral Experience, Vall d'Hebron Institute of Oncology (VHIO)
2018-current
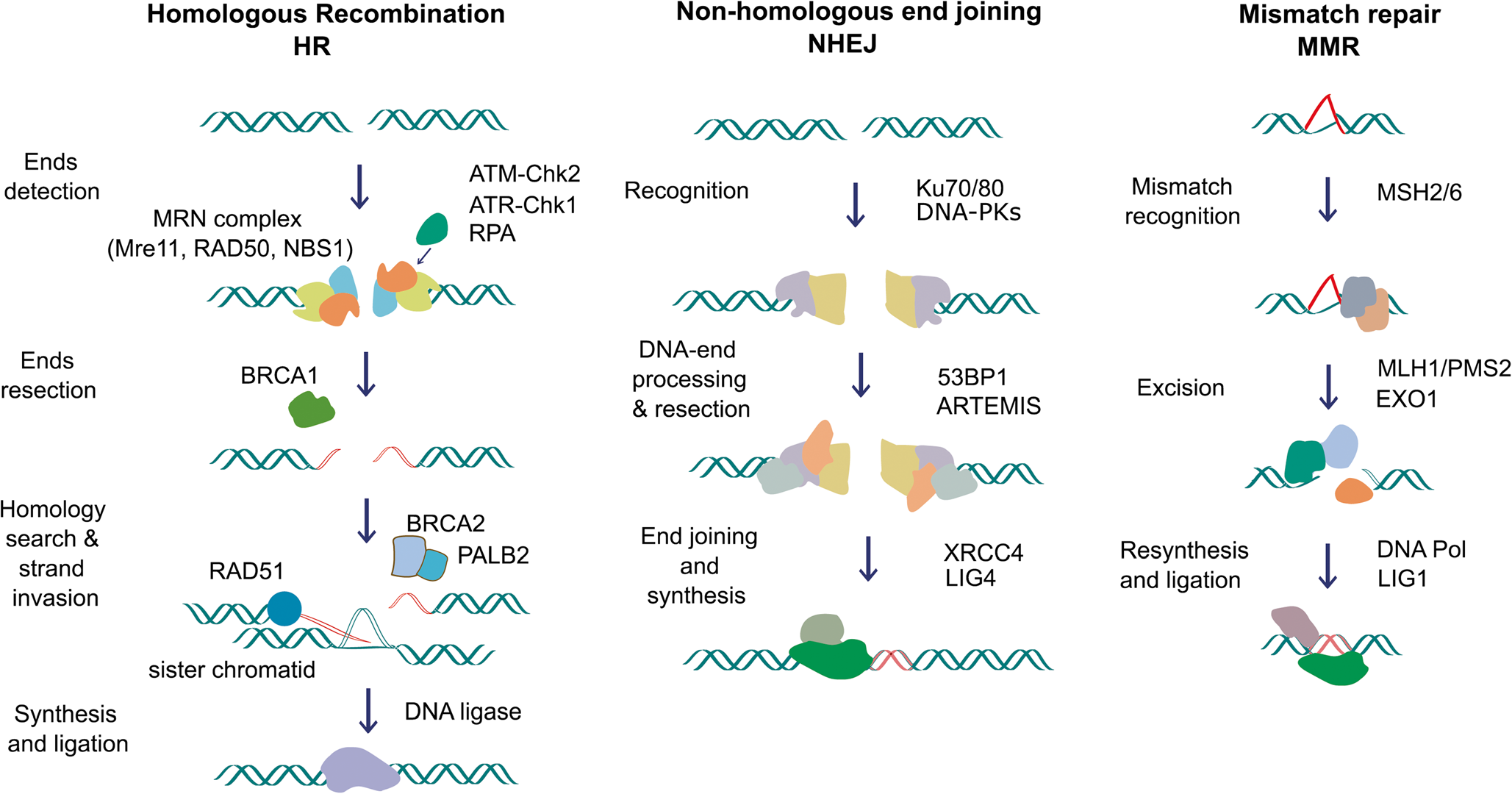
Prostate Cancer Research: I am currently working in a project to identify copy number profiles signatures in Whole Genomes associated with alterations in DNA Damage Response (DDR) genes. In addition I am analyzing genomics and transcriptomic data from liquid biopsy samples.
Graduate student, Center for Applied Medical Research (CIMA)
2012-2017
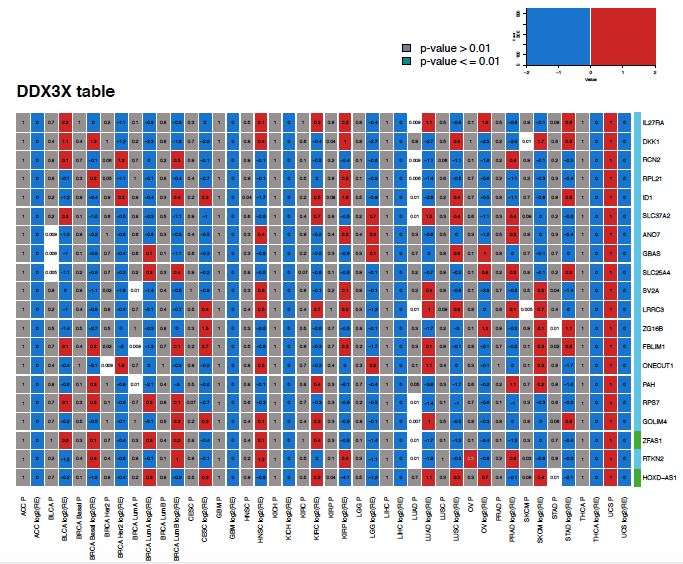
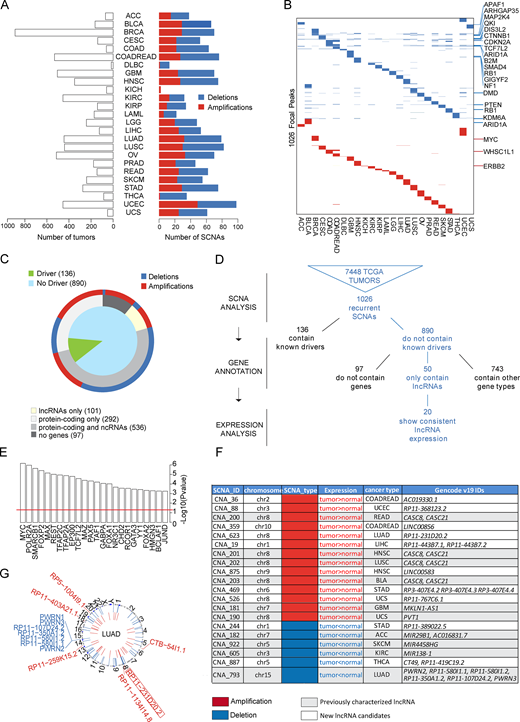
Integrative Genomic Analysis: Analyzed the genome of 7,448 tumors from cancer patients to identify copy number altered regions harboring a class of genes called: lncRNAs. Integrated these data with RNA-seq to identify novel cancer genes. In addition, experimentally validated the function of a discovered lncRNA and confirmed its roles in lung cancer.
Analysis of Cancer Mutations: Identified the mutations present in cancer tumors samples and analyzed their impact in RNA binding proteins (RBPs). In addition these data was integrated with eCLIP and shRNAs experiments to unveil RBPs-RNA interactions involved in the disease.
Master student, Herbert Irving Comprehensive Cancer Center (HICC)
2010-2011
Breast Cancer Research: Experimentally explored the regulation of the tumor suppressor PTEN performing molecular biology techniques.
Emerging Roles of the Sirtuin Family: I wrote the academic review where I described the key aspects of sirtuin's biology, presents the current situation of sirtuins in the biotechnology industry, an its function in cell metabolism, chromatin regulation and cancer. This review is available at Columbia University Academic Commons: https://doi.org/10.7916/D8X354FT.
Rotation student, Institute of Cellular Physiology (IFC)
2006-2010
Influenza Virus Detection: Conducted sequence comparison analysis for the design of molecular beacon probes to differentially detect Influenza virus subtypes during the global flu outbreak in 2009.
Education
- University of Navarra: PhD in Biomedicine (2012-2017) Pamplona, Spain.
- Columbia University: MA in Biotechnology (2010-2011) New York, USA.
- National University of México: BS in Genomic Sciences (2006-2010) DF, México.